Our Assay
Overview
We have developed an in-vitro assay to test different guide RNAs across a range of conditions (e.g. temperature) for both Cas9 and Cas12a nucleases to help choose guide RNAs that are efficient under different experimental growth conditions.
We can take into account the GC content of your genome, specific regions you want to target, editing approach (single or multiple targets) and preferred nuclease to help you tailor genome editing to your species and experimental set-up.
For example, Cas9 is currently the most popular nuclease for genome editing. Whilst both Cas9 and Cas12a can demonstrate activity below the optimum of 37°C, Cas12a can maintain activity at lower temperatures. Differences in the protospacer adjacent motif (PAM) sequence for each nuclease (NGG for Cas9 and TTTV for Cas12a) means that target site and number can vary for each nuclease across genomes and genes.
Our service helps you design your CRISPR-Cas experiments and tests your preferred guide RNAs to identify those that cut with the highest efficiency in-vitro. This enables you to make an informed choice of guide RNA before moving onto often timely and costly in-vivo work.
There are many factors which can affect and delay the ability to carry out successful genome editing, such as transformation method, selection of potential mutants and the ability of your species to induce mutations through DNA repair mechanisms following cutting by a CRISPR-Cas nuclease. There are also physical barriers within the genome such as the presence of heterochromatin which can reduce accessibility of the CRISPR-Cas complex.
Whilst you work hard to overcome these aspects of your method, we can help to design your guide RNAs and test them in-vitro for you to make sure you are targeting regions which have the potential to cut efficiently under your experimental conditions.
- We combine carefully designed guide RNA's, your choice of CRISPR nuclease and your target sequence under conditions relevant to your experimental set-up, to screen guide RNA's for cutting efficiency before you carry out labour/time intensive in-vivo editing. We can also send you the tested guide RNA's for use in editing.
- We have used the assay to successfully pre-screen guide RNA's for several different species and genes to identify guide RNA's that are active and efficient under our growth conditions.
- Our assay can be adapted for other CRISPR nucleases. Please contact us if you are interested in using a different nuclease.
In vitro assay with CPF1
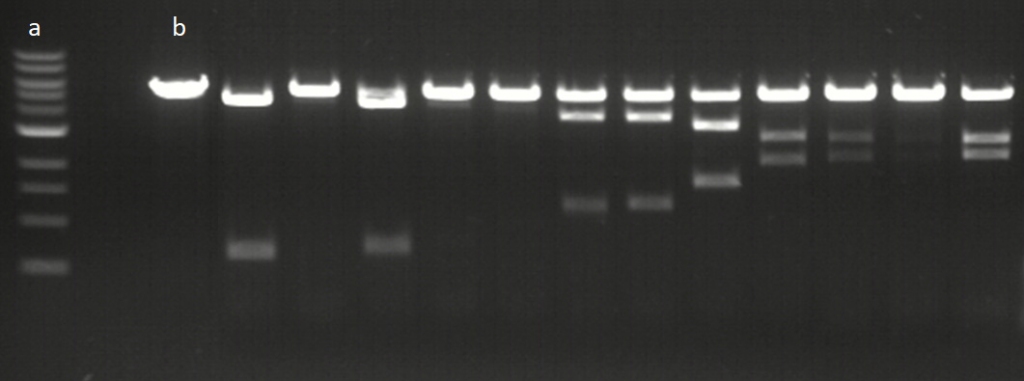
Example of assay results for 12 Cas12a gRNAs at 20°C. a; ladder, b; control plasmid without editing. Editing efficiencies vary between target sequences with a single band representing no editing, 3 bands showing degrees of partial editing and 2 bands showing efficient editing to completion.

